
Getting Started with geospatialsuite
geospatialsuite Development Team
Source:vignettes/getting-started.Rmd
getting-started.RmdIntroduction
geospatialsuite is a comprehensive R package for geospatial analysis and visualization. It provides universal functions that work with any region, robust error handling, and simplified workflows for complex spatial analysis tasks.
Key Features
- 60+ vegetation indices with automatic band detection
- Universal spatial mapping with one-line functionality
- Reliable terra-based visualization without complex dependencies
- Agricultural applications including CDL crop analysis
- Water quality assessment with multiple indices
- Robust error handling throughout all functions
- Built-in sample data for learning and testing
- Flexible data loading from files, directories, or objects
Installation
# Install from CRAN
install.packages("geospatialsuite")
# Load the package
library(geospatialsuite)
library(terra)Quick Start with Built-in Sample Data
geospatialsuite includes built-in sample data so you can start immediately:
library(geospatialsuite)
library(terra)
# List available sample datasets
list_sample_datasets()
#> filename type size_kb
#> 1 sample_red.rds SpatRaster 2
#> 2 sample_nir.rds SpatRaster 2
#> 3 sample_blue.rds SpatRaster 2
#> 4 sample_green.rds SpatRaster 2
#> 5 sample_swir1.rds SpatRaster 2
#> 6 sample_multiband.rds SpatRaster 8
#> 7 sample_points.rds sf 3
#> 8 sample_boundary.rds sf 2
#> 9 sample_coordinates.csv data.frame 1
#> description
#> 1 Red band reflectance (10x10 pixels, Ohio region)
#> 2 NIR band reflectance (10x10 pixels, Ohio region)
#> 3 Blue band reflectance (10x10 pixels, Ohio region)
#> 4 Green band reflectance (10x10 pixels, Ohio region)
#> 5 SWIR1 band reflectance (10x10 pixels, Ohio region)
#> 6 Multi-band raster (Blue, Green, Red, NIR, SWIR1)
#> 7 Sample field locations (20 points with attributes)
#> 8 Sample study area boundary polygon
#> 9 Sample coordinates with elevation and soil data
#> use_case available
#> 1 Vegetation index calculation (NDVI, SAVI, etc.) TRUE
#> 2 Vegetation index calculation (NDVI, SAVI, etc.) TRUE
#> 3 Enhanced vegetation index (EVI) calculation TRUE
#> 4 Water index calculation (NDWI, GNDVI) TRUE
#> 5 Water and moisture indices (NDMI, MNDWI) TRUE
#> 6 Auto band detection, multi-index calculation TRUE
#> 7 Spatial join examples, extraction workflows TRUE
#> 8 Region boundary examples, masking operations TRUE
#> 9 Geocoding, spatial integration examples TRUE
#> access_method
#> 1 load_sample_data('sample_red.rds')
#> 2 load_sample_data('sample_nir.rds')
#> 3 load_sample_data('sample_blue.rds')
#> 4 load_sample_data('sample_green.rds')
#> 5 load_sample_data('sample_swir1.rds')
#> 6 load_sample_data('sample_multiband.rds')
#> 7 load_sample_data('sample_points.rds')
#> 8 load_sample_data('sample_boundary.rds')
#> 9 load_sample_data('sample_coordinates.csv')
# Load sample raster data
red <- load_sample_data("sample_red.rds")
nir <- load_sample_data("sample_nir.rds")
# Calculate NDVI
ndvi <- calculate_vegetation_index(red = red, nir = nir, index_type = "NDVI")
# Visualize
plot(ndvi, main = "NDVI from Sample Data", col = terrain.colors(100))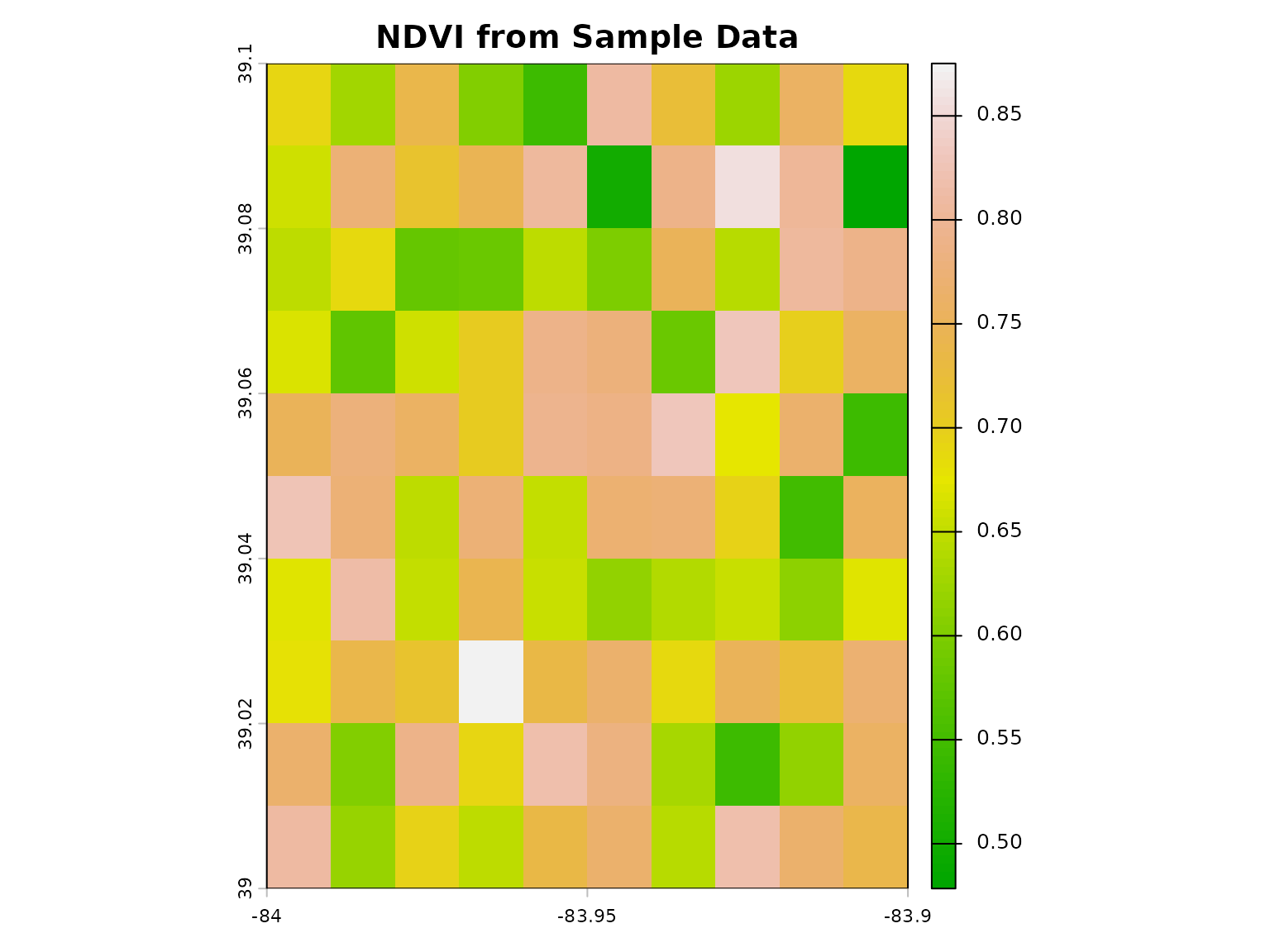
Core Workflows
1. Calculate Vegetation Indices
# Load sample spectral bands
red <- load_sample_data("sample_red.rds")
nir <- load_sample_data("sample_nir.rds")
blue <- load_sample_data("sample_blue.rds")
# Calculate NDVI
ndvi <- calculate_vegetation_index(red = red, nir = nir, index_type = "NDVI")
plot(ndvi, main = "NDVI")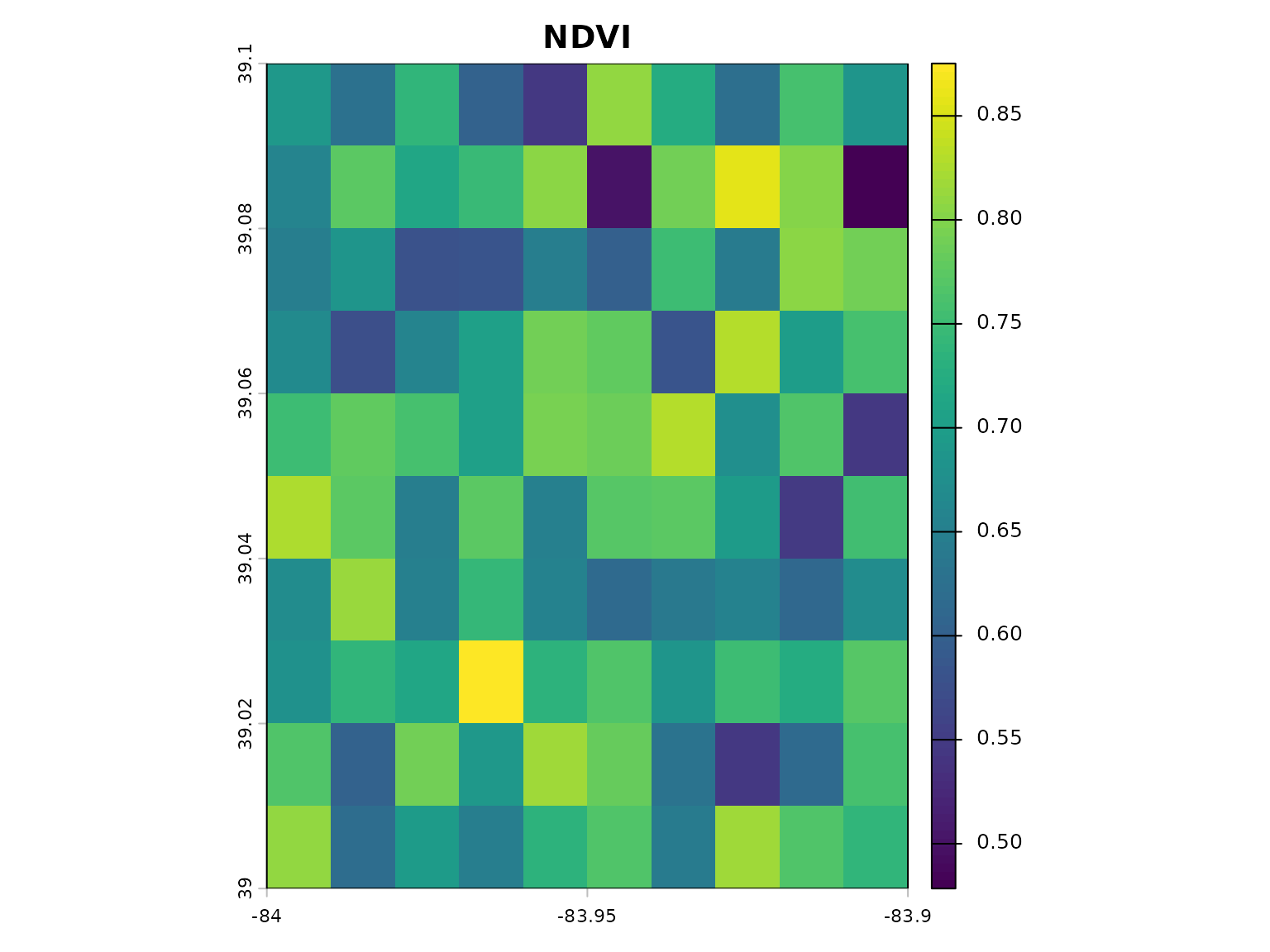
# Calculate EVI (requires blue band)
evi <- calculate_vegetation_index(red = red, nir = nir, blue = blue, index_type = "EVI")
plot(evi, main = "EVI")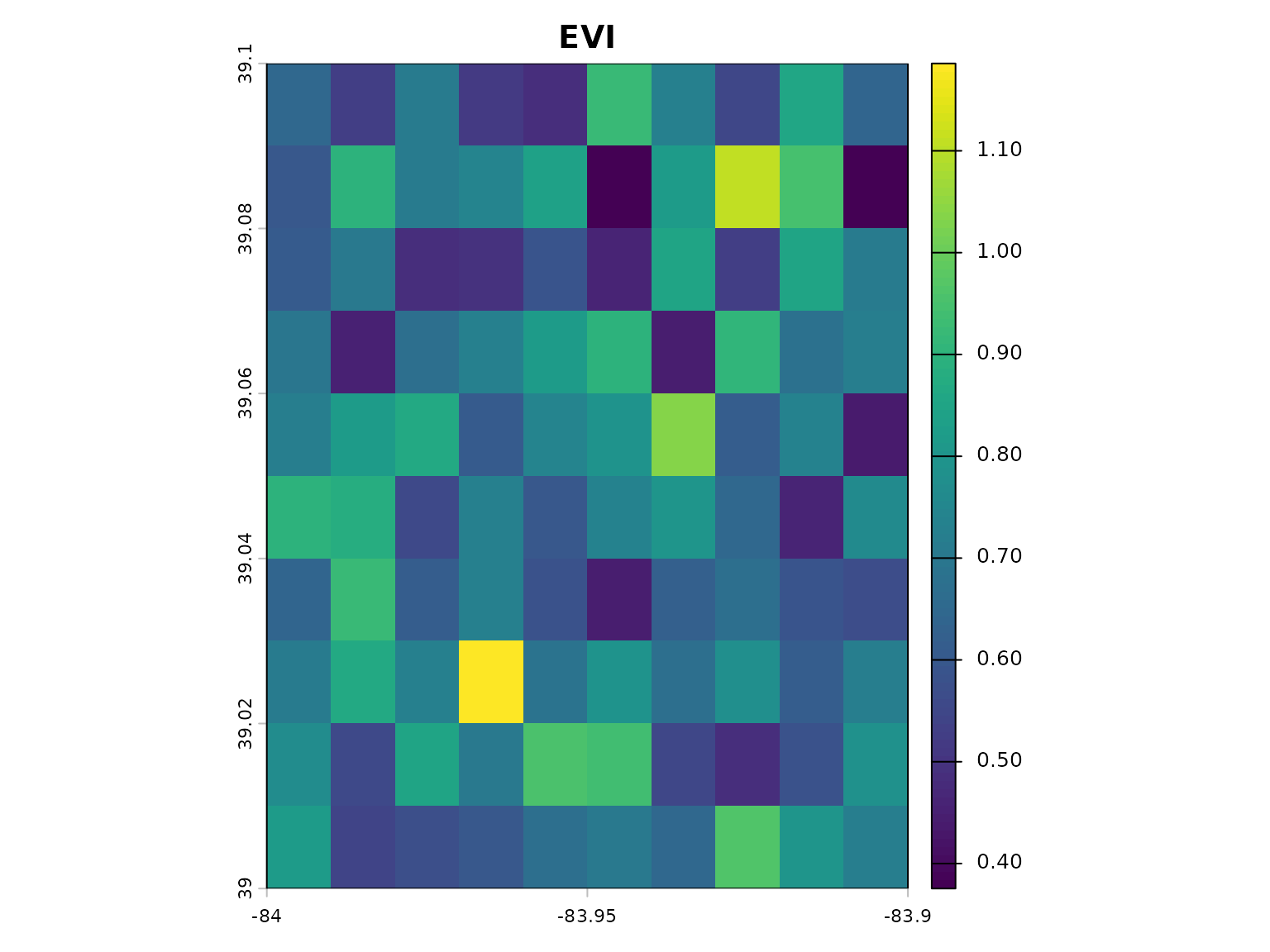
# Calculate multiple indices at once
indices <- calculate_multiple_indices(
red = red,
nir = nir,
blue = blue,
indices = c("NDVI", "EVI", "SAVI"),
output_stack = TRUE
)
# Plot all indices
plot(indices)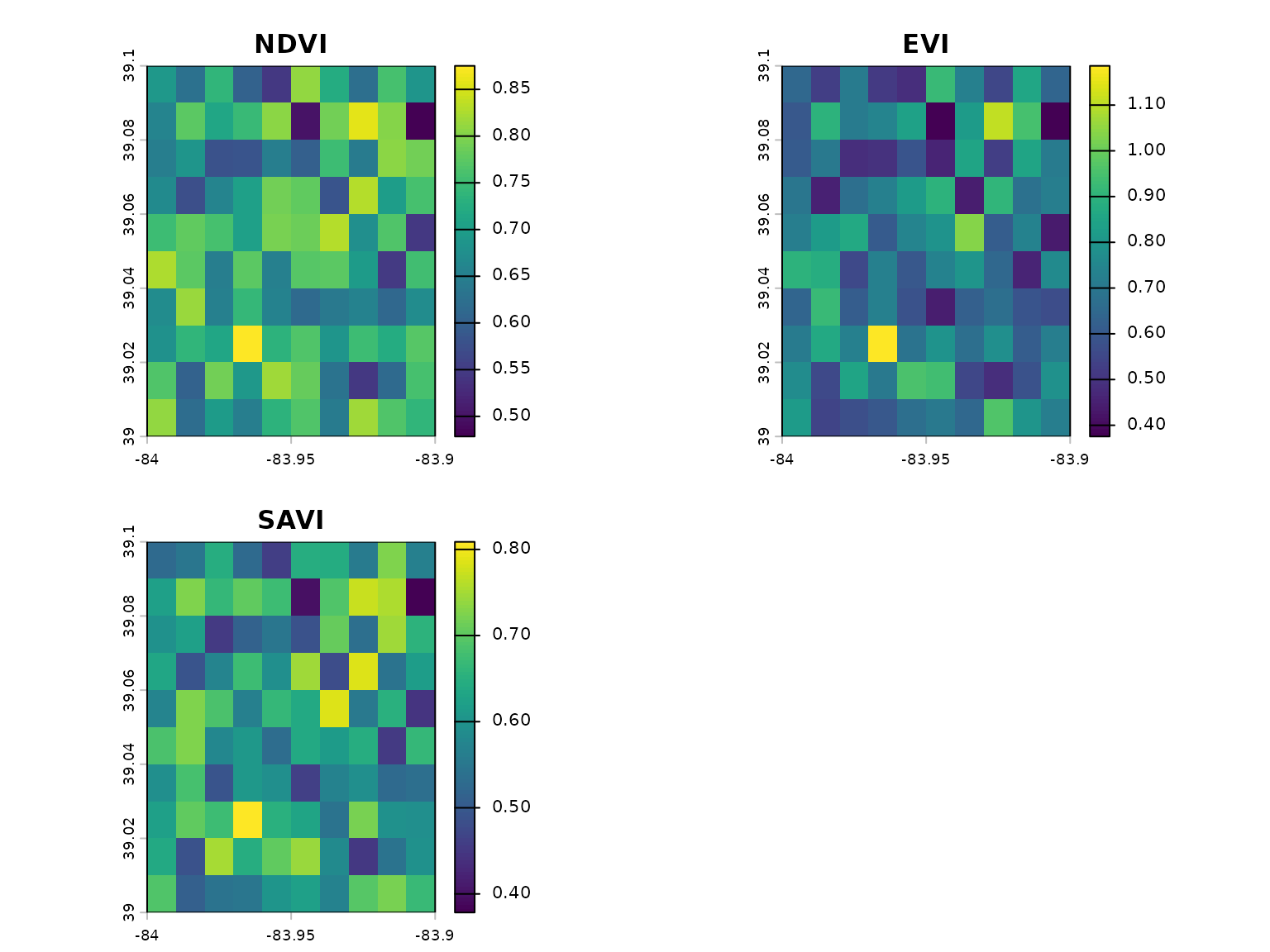
2. Working with Multi-band Data
# Load multi-band sample raster
multiband <- load_sample_data("sample_multiband.rds")
# Check band names
names(multiband)
#> [1] "blue" "green" "red" "nir" "swir1"
# Auto-detect bands and calculate index
ndvi_auto <- calculate_vegetation_index(
spectral_data = multiband,
index_type = "NDVI",
auto_detect_bands = TRUE
)3. Spatial Operations
# Load sample vector data
points <- load_sample_data("sample_points.rds")
boundary <- load_sample_data("sample_boundary.rds")
# Calculate NDVI
red <- load_sample_data("sample_red.rds")
nir <- load_sample_data("sample_nir.rds")
ndvi <- calculate_vegetation_index(red = red, nir = nir, index_type = "NDVI")
# Extract raster values to points
points_with_values <- universal_spatial_join(
source_data = points,
target_data = ndvi,
method = "extract"
)
# Check result
head(points_with_values)
#> Simple feature collection with 6 features and 7 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -83.94662 ymin: 39.00987 xmax: -83.91607 ymax: 39.09223
#> Geodetic CRS: WGS 84
#> site_id ndvi evi crop_type elevation_m soil_moisture
#> 1 SITE_01 0.593 0.274 soybeans 269 0.27
#> 2 SITE_02 0.660 0.485 wheat 283 0.28
#> 3 SITE_03 0.646 0.579 corn 211 0.21
#> 4 SITE_04 0.388 0.483 pasture 256 0.31
#> 5 SITE_05 0.776 0.588 corn 219 0.35
#> 6 SITE_06 0.645 0.539 soybeans 205 0.30
#> geometry extracted_NDVI
#> 1 POINT (-83.94662 39.04604) 0.7709993
#> 2 POINT (-83.94162 39.00987) 0.7667765
#> 3 POINT (-83.94104 39.02066) 0.7647660
#> 4 POINT (-83.9309 39.09223) 0.7207893
#> 5 POINT (-83.93327 39.03194) 0.6405917
#> 6 POINT (-83.91607 39.02653) 0.72155344. Data Visualization
# Load sample data
points <- load_sample_data("sample_points.rds")
# Quick map (auto-detects everything)
quick_map(points, variable = "ndvi", title = "NDVI at Sample Sites")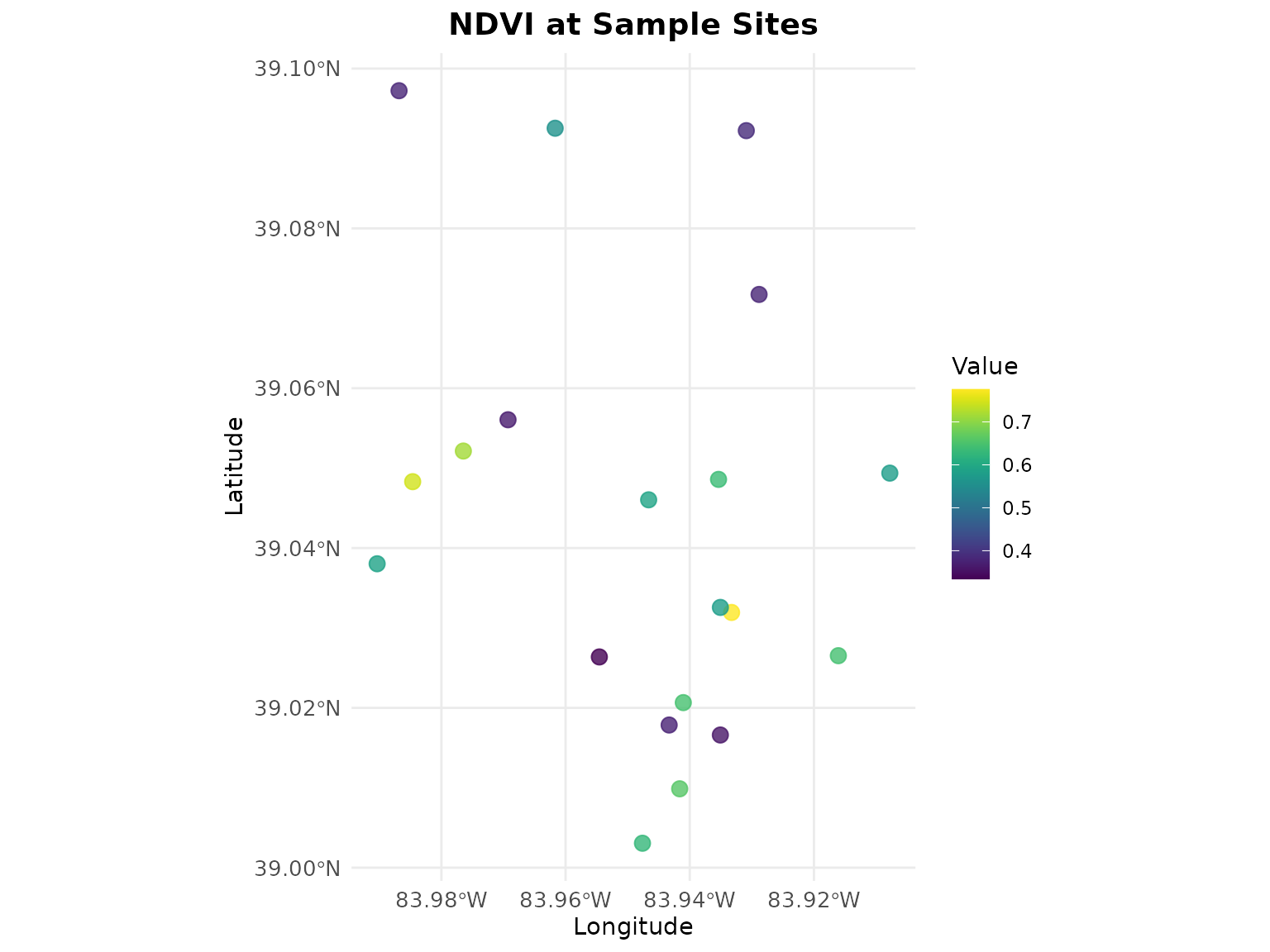
Available Sample Datasets
# List all available datasets
datasets <- list_sample_datasets()
print(datasets[, c("filename", "type", "description")])
#> filename type
#> 1 sample_red.rds SpatRaster
#> 2 sample_nir.rds SpatRaster
#> 3 sample_blue.rds SpatRaster
#> 4 sample_green.rds SpatRaster
#> 5 sample_swir1.rds SpatRaster
#> 6 sample_multiband.rds SpatRaster
#> 7 sample_points.rds sf
#> 8 sample_boundary.rds sf
#> 9 sample_coordinates.csv data.frame
#> description
#> 1 Red band reflectance (10x10 pixels, Ohio region)
#> 2 NIR band reflectance (10x10 pixels, Ohio region)
#> 3 Blue band reflectance (10x10 pixels, Ohio region)
#> 4 Green band reflectance (10x10 pixels, Ohio region)
#> 5 SWIR1 band reflectance (10x10 pixels, Ohio region)
#> 6 Multi-band raster (Blue, Green, Red, NIR, SWIR1)
#> 7 Sample field locations (20 points with attributes)
#> 8 Sample study area boundary polygon
#> 9 Sample coordinates with elevation and soil dataSample datasets include:
-
sample_red.rds- Red band SpatRaster (10×10 pixels) -
sample_nir.rds- NIR band SpatRaster -
sample_blue.rds- Blue band SpatRaster -
sample_green.rds- Green band SpatRaster -
sample_swir1.rds- SWIR1 band SpatRaster -
sample_multiband.rds- Multi-band stack (5 bands) -
sample_points.rds- Sample field locations (sf object) -
sample_boundary.rds- Study area polygon (sf object) -
sample_coordinates.csv- Tabular data with coordinates
Working with Your Own Data
Loading Raster Files with geospatialsuite
Single GeoTIFF File
# Use geospatialsuite's load_raster_data() function
# It provides robust error handling and validation
# Load a single .tif file
my_raster <- load_raster_data("path/to/your/ndvi.tif")
# Result is a list, extract the raster
ndvi_raster <- my_raster[[1]]
# Now use with geospatialsuite functions
# The raster is ready for analysis
summary(ndvi_raster)Multiple GeoTIFF Files
# Load multiple Landsat bands with geospatialsuite
# Handles validation automatically
landsat_files <- c(
"LC08_B4_red.tif",
"LC08_B5_nir.tif",
"LC08_B3_green.tif"
)
# geospatialsuite loads them with error checking
bands <- load_raster_data(landsat_files, verbose = TRUE)
# Extract individual bands
red_band <- bands[[1]]
nir_band <- bands[[2]]
green_band <- bands[[3]]
# Calculate indices using geospatialsuite
ndvi <- calculate_vegetation_index(
red = red_band,
nir = nir_band,
index_type = "NDVI"
)
gndvi <- calculate_vegetation_index(
green = green_band,
nir = nir_band,
index_type = "GNDVI"
)Load from Directory
# geospatialsuite can load all rasters from a directory
# Perfect for batch processing
# Load all .tif files from Landsat directory
all_bands <- load_raster_data(
"/path/to/landsat/imagery/",
pattern = "\\.(tif|tiff)$",
verbose = TRUE
)
# geospatialsuite finds, loads, and validates all files
# Returns a list of SpatRaster objects ready to use
cat("Loaded", length(all_bands), "raster files\n")Real-World Landsat Workflow
# Complete workflow using geospatialsuite functions
library(geospatialsuite)
# 1. Load Landsat bands using geospatialsuite
landsat_bands <- load_raster_data(
"landsat/LC08_L2SP_021033_20240715/",
pattern = "SR_B[2-5].TIF$",
verbose = TRUE
)
# geospatialsuite loaded them with validation
# Extract bands (scaled values 0-1 after Collection 2 scaling)
blue <- landsat_bands[[1]] # After scaling
green <- landsat_bands[[2]]
red <- landsat_bands[[3]]
nir <- landsat_bands[[4]]
# 2. Calculate vegetation indices using geospatialsuite
# The package has 60+ pre-programmed indices
veg_indices <- calculate_multiple_indices(
red = red,
nir = nir,
blue = blue,
green = green,
indices = c("NDVI", "EVI", "SAVI", "GNDVI"),
output_stack = TRUE
)
# 3. Visualize using geospatialsuite
quick_map(veg_indices$NDVI, title = "Landsat 8 NDVI - July 15, 2024")Working with Vector Data
Loading Shapefiles
# Load shapefile with sf (standard approach)
library(sf)
field_boundaries <- sf::st_read("data/farm_fields.shp")
# Then use with geospatialsuite's spatial functions
# Calculate NDVI first
ndvi <- calculate_vegetation_index(red = red, nir = nir, index_type = "NDVI")
# Extract NDVI to field boundaries using geospatialsuite
fields_with_ndvi <- universal_spatial_join(
source_data = field_boundaries,
target_data = ndvi,
method = "extract"
)
# geospatialsuite handles CRS mismatches automatically
# Returns field boundaries with extracted NDVI statistics
head(fields_with_ndvi)Working with GeoPackages
# Load GeoPackage (modern format, better than shapefile)
farm_data <- sf::st_read("farm_management.gpkg", layer = "fields")
sample_points <- sf::st_read("farm_management.gpkg", layer = "samples")
# Use geospatialsuite's spatial join
samples_with_indices <- universal_spatial_join(
source_data = sample_points,
target_data = veg_indices,
method = "extract",
buffer_distance = 30 # 30m buffer
)
# geospatialsuite extracted all indices in the stack
# Each index becomes a column in the result
names(samples_with_indices)Multi-band Raster with Auto-Detection
# geospatialsuite's auto-detection feature
# Load a stacked multi-band GeoTIFF
multiband_raster <- load_raster_data("sentinel2_stack.tif")[[1]]
# Name the bands (Sentinel-2 example)
names(multiband_raster) <- c("blue", "green", "red", "nir", "swir1")
# Use geospatialsuite's auto-detection
# It finds the right bands automatically!
indices <- calculate_multiple_indices(
spectral_data = multiband_raster,
indices = c("NDVI", "EVI", "MNDWI"),
auto_detect_bands = TRUE, # This is geospatialsuite's feature!
output_stack = TRUE
)
# No need to specify which band is which
# geospatialsuite figured it out!Complete Real-World Example
# End-to-end agricultural monitoring with geospatialsuite
library(geospatialsuite)
library(sf)
# 1. Load satellite imagery using geospatialsuite
spectral_bands <- load_raster_data(
"/path/to/satellite/bands/",
pattern = "B[0-9].tif$",
verbose = TRUE
)
# 2. Extract bands
red <- spectral_bands[[3]]
nir <- spectral_bands[[4]]
green <- spectral_bands[[2]]
# 3. Calculate indices using geospatialsuite
crop_health <- calculate_multiple_indices(
red = red,
nir = nir,
green = green,
indices = c("NDVI", "GNDVI", "SAVI"),
output_stack = TRUE
)
# 4. Load field data
fields <- sf::st_read("farm_data/fields.shp")
# 5. Extract to fields using geospatialsuite
fields_analysis <- universal_spatial_join(
source_data = fields,
target_data = crop_health,
method = "extract"
)
# 6. Visualize using geospatialsuite
quick_map(fields_analysis,
variable = "NDVI",
title = "Field Health Assessment")
# geospatialsuite handled:
# - Loading multiple files
# - Calculating indices
# - Spatial extraction with CRS handling
# - VisualizationListing Available Indices
# See all vegetation indices geospatialsuite provides
veg_indices <- list_vegetation_indices()
head(veg_indices[, c("Index", "Category", "Description")])
#> Index Category Description
#> 1 NDVI basic Normalized Difference Vegetation Index
#> 2 SAVI basic Soil Adjusted Vegetation Index
#> 3 MSAVI basic Modified Soil Adjusted Vegetation Index
#> 4 OSAVI basic Optimized Soil Adjusted Vegetation Index
#> 5 EVI basic Enhanced Vegetation Index
#> 6 EVI2 basic Two-band Enhanced Vegetation Index
# See water indices
water_indices <- list_water_indices()
head(water_indices)
#> Index Type Required_Bands Primary_Application
#> 1 NDWI Water Detection Green, NIR water_detection
#> 2 MNDWI Water Detection Green, SWIR1 water_detection
#> 3 NDMI Vegetation Moisture NIR, SWIR1 moisture_monitoring
#> 4 MSI Moisture Stress NIR, SWIR1 drought_assessment
#> 5 NDII Vegetation Moisture NIR, SWIR1 moisture_monitoring
#> 6 WI Water Content NIR, SWIR1 moisture_monitoring
#> Description
#> 1 Normalized Difference Water Index (McFeeters 1996) - Original water detection index
#> 2 Modified NDWI (Xu 2006) - Enhanced water detection, reduces built-up area confusion
#> 3 Normalized Difference Moisture Index (Gao 1996) - Vegetation water content
#> 4 Moisture Stress Index - Plant water stress detection (lower = more moisture)
#> 5 Normalized Difference Infrared Index - Alternative name for vegetation moisture
#> 6 Water Index - Simple ratio for water content assessment
#> Value_Range Water_Threshold Reference
#> 1 [-1, 1] > 0.3 McFeeters (1996)
#> 2 [-1, 1] > 0.5 Xu (2006)
#> 3 [-1, 1] N/A (vegetation) Gao (1996)
#> 4 [0, 10+] < 1.0 Various
#> 5 [-1, 1] N/A (vegetation) Hunt & Rock (1989)
#> 6 [0, 10+] > 1.0 VariousGetting Help
# Package documentation
help(package = "geospatialsuite")
# Function help
?calculate_vegetation_index
?load_raster_data
?universal_spatial_join
?quick_map
# Test package installation
test_geospatialsuite_package_simple()
#> $test_results
#> $test_results$basic_ndvi_test
#> [1] TRUE
#>
#> $test_results$water_index_test
#> [1] TRUE
#>
#> $test_results$basic_visualization_test
#> [1] TRUE
#>
#> $test_results$multiple_indices_simple_test
#> [1] TRUE
#>
#> $test_results$enhanced_ndvi_simple_test
#> [1] TRUE
#>
#> $test_results$dependencies_test
#> [1] TRUE
#>
#> $test_results$spatial_operations_test
#> [1] TRUE
#>
#> $test_results$data_loading_test
#> [1] TRUE
#>
#>
#> $summary
#> $summary$total_tests
#> [1] 8
#>
#> $summary$passed_tests
#> [1] 8
#>
#> $summary$failed_tests
#> [1] 0
#>
#> $summary$success_rate
#> [1] 100
#>
#> $summary$duration_seconds
#> [1] 0.22
#>
#> $summary$version
#> [1] "0.1.0"
#>
#>
#> $test_output_dir
#> [1] "/tmp/RtmpDaZ1bR"
#>
#> $timestamp
#> [1] "2026-02-09 15:50:24 UTC"
#>
#> $test_approach
#> [1] "simplified_robust"
#>
#> $core_message
#> [1] "Focused on essential functionality with minimal complexity"Summary
geospatialsuite provides:
Data Loading:
-
load_raster_data()- Load .tif files with validation -
load_sample_data()- Access built-in samples
Analysis:
-
calculate_vegetation_index()- 60+ indices -
calculate_multiple_indices()- Batch processing - Auto band detection feature
Spatial Operations:
-
universal_spatial_join()- Extract values, automatic CRS handling
Visualization:
-
quick_map()- One-line mapping -
create_spatial_map()- Custom maps
All with robust error handling and simplified workflows!
For more detailed tutorials, see the other vignettes:
- Vegetation Indices Analysis
- Agricultural Applications
- Spatial Analysis and Integration
- Water Quality Assessment
- Complete Workflows and Case Studies